Class 12 VBQs Biology Molecular Basis of Inheritance
Short Answer Type Questions
Question. (i) Why did Hershey and Chase use radioactivesulfur and radioactive phosphorus in their experiment ?
(ii) Write the conclusion they arrived at and how.
Answer : (i) Sulphur is a component of protein and thus would label the protein coat. Phosphorus is component of DNA.
(ii) Bacteria which were infected with viruses having radioactive DNA were found to contain radioactive DNA later on.
Bacteria which were infected with viruses having radioactive protein coat were not found to contain radioactivity.
Conclusion-DNA is the genetic material.
Question. (i) Differentiate between a template strand and coding strand of DNA.
(ii) Name the source of energy for the replication of DNA.
Answer :
| (i) Role/Strand | Template strand | Coding strand |
| Function | Codes for the protein molecule | Does not code for anything |
| Polarity | 3’ → 5’ | 5’ → 3’ |
(ii) Deoxynucleoside triphosphates.
Question. (i) A DNA segment has a total of 1000 nucleotides, out of which 240 of them are adenine containing nucleotides. How many pyrimidine bases this DNA segment possesses ?
(ii) Draw a diagrammatic sketch of a portion of DNA segment to support your answer.
Answer : (i) (a) A = T, A = 240 hence T = 240
A + T = 240 + 240 = 480
So, G + C = 1000 – 480 = 520
G = C, so C = 520/2 = 260
So, pyrimidines = C + T
= 260 + 240 = 500
(b) Purine A and G always pair with T and C respectively
(c) A/G =T/C =1 (Chargaff rule)
(ii)


Diagram showing polarity = ½
A – T = ½
G – C = ½
H-bond = ½ 1+2
Detailed Answer :
(i) Adenine and guanine are purines. Cytosine and thymine are pyrimidines. According to Chargaff’s complementarity rule, the amount of purines is always equal to amount of pyrimidines.
Thus, the numbers of adenine (A) will be equal to the number of thymine (T).
Number of Adenine (A) containing nucleotides = 240
Thus, A = T = 240
Therefore, A + T = 240 + 240 = 480
The number of cytosine (C) will be equal to number of guanine (G).
Thus, G + C = Total number of nucleotides.
Nucleotides containing A and T nitrogenous
bases= 1000 – 480 = 520
Therefore, G = 260, C = 260
Number of guanine will be equal to number of cytosine which will be 260.
Therefore, the number of pyrimidines that the segment possess = C + T = 260 + 240 = 500
Question. (i) Write what DNA replication refers to.
(ii) State the properties of DNA replication model.
(iii) List any three enzymes involved in the process along with their functions.
Answer : (i) DNA synthesis i.e. copying of DNA
(ii) (a) Semi-conservative.
(b) Semi-discontinuous.
(c) Unidirectional.
(iii) DNA polymerase III – adds nucleotides.
DNA polymerase I – fills the gaps.
RNA primase – brings primers.
Topoisomerase – causes unwinding.
DNA ligase – joins Okazaki fragments.
Question. Draw a labelled diagram of nucleosome. Where is it found in a cell ?
Answer :

Label any three parts
Location : Chromatin of nucleus.
Question. Describe the experiments that established the identity of ‘transforming principles‘ of Griffith.
Answer : (i) Purification of biochemicals like Proteins, RNA & DNA from S cells (heat killed).
(ii) Presence of Protein & RNA in medium did not affect transformation.
(iii) DNA alone from S Bacteria caused R Bacteria to transform.
(iv) Digestion with DNAase inhibits transformation.
Conclusion : DNA is the transforming chemical / biochemical material as it was transferred from virus to bacteria.
Detailed Answer :
In 1944, Avery MacLeod and McCarty worked to determine the chemical nature of transforming principle‘.
They purified biochemicals from heat killed S-cells.
(i) Proteins : Proteases–Transformation takes place So, protein is not a ‘transforming principle‘.
(ii) RNA : RNases–Transformation takes place. So, RNA is not a ‘transforming Principle‘.
(iii) DNA : DNases–Transformation inhibited. Therefore, DNA is the ‘Transforming Principle‘.
Question. Explain the role of DNA-dependent RNA polymerase in transcription.
Answer : RNA polymerase catalyzes RNA synthesis. To transcribe a gene, RNA polymerase proceeds through a series of well-defined steps, which are grouped into three phases : initiation, elongation and termination.
(i) Initiation : RNA polymerase binds with the promoter region of the DNA. The promoter – polymerase complex undergoes structural changes required for transcription.
(ii) Elongation : During elongation, RNA polymerase unwinds the DNA in front and re-anneals it behind, it dissociates the growing RNA chain from the template as it moves along.
It also performs proof reading function.
(iii) Termination : Once the polymerase has transcribed the length of the gene, it stops and releases the RNA product.
Question. (i) Draw a neat labelled diagram of a nucleosome.
(ii) Mention what enables histones to acquire a positive charge.
Answer : (i)

(ii) Basic amino acid residues of lysines, arginines.
Detailed Answer :
(i) Histones acquire positive charge because they are rich in basic amino acid residues of arginine and lysine, which carry positive charges in their side chains.
Question. The base sequence in one of the strands of DNA is TAGCATGAT.
(i) Give the base sequence of its complementary strand.
(ii) How are these base pairs held together in a DNA molecule ?
(iii) Explain the base complementarity rules. Name the scientist who framed this rule.
Answer : (i) ATCGTACTA
(ii) Through Hydrogen bonds between A and T and C and G on the two strands.
(iii) A = T and C → G, Watson and Crick / Chargaff.
Detailed Answer :
(i) ATCGTACTA.
(ii) The base pairs are held together in a DNA molecule by hydrogen bond in such a way that Adenine (A) pairs with Thymine (T) by two Hydrogen bonds (A = T) and Guanine pairs with Cytosine (C) by three Hydrogen bonds (C → G).
(iii) The base complementarity rule is that the ratios between adenine-thymine and guanine-cytosine are constant and equal to one. The scientists who framed this rule were Erwin Chargaff and Watson and Crick.
Question. Identify A, B, C, D, E and F in the following table.
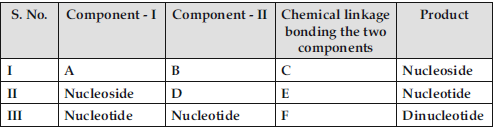
Answer : (I) A – Nitrogenous base / A – Pentose sugar.
B – Pentose Sugar / B – Nitrogenous base.
C – N-glycosidic linkage.
(II) D – phosphate group.
E – phospho ester linkage.
(III) F – (3′ – 5′) phosphodiester linkage.
Question. In a typical nucleus, some regions of chromatin are stained light and others dark. Explain why is it so and what is its significance ?
Answer : Light stained-loosely packed-Euchromatin.
Dark stained-densely packed-Heterochromatin.
Euchromatin-transcriptionally active.
Heterochromatin-transcriptionally inactive.
Detailed Answer :
Euchromatin is lightly stained, diffused but narrow fibrous part of chromatin. It is normal chromatin which possesses active genes. It is transcriptionally active. During nuclear division it replicates normally. It is formed through loose spiralization of nucleosome strands.
Heterochromatin is darkly stained granular part of chromatin and is formed by solenoid type of coiling of nucleosomes. Transcription does not occur in these regions. Active genes are also absent in it.
Question. Name the specific enzyme responsible for nucleotide polymerisation in DNA replication.
Write two characteristic features of this enzyme.
Name the region on E. coli DNA where this enzyme can initiate replication.
Answer : (i) DNA – dependent DNA polymerase.
(ii) The enzyme uses DNA template to catalyse the polymerisation of deoxyribonucleotides.
(iii) Have to catalyse the reaction with high accuracy.
(iv) Origin of replication / Ori.
Question. Name the three RNA polymerases found in eukaryotic cells and mention their functions.
Answer : RNA polymerase – I, transcribes rRNAs (28S -18S and 5.8S).
RNA polymerase – II, transcribes precursor of mRNA / hnRNA / heterogeneous RNA.
RNA polymerase – III, transcribes tRNA / 5Sr RNA / snRNA.
Question. A DNA segment has a total of 1500 nucleotides out of which 410 are guanine containing nucleotides.
How many Pyrimidine bases this DNA segment possesses.
Answer : ∴ G = 410 ∴C = 410 (because C = G)
∴ G + C = 410 + 410 = 820
∴ A + T = 1500 – 820 = 680
∴ T = 680/ 2 = 340
Thus the pyrimidine bases C + T
= 410 + 340
= 750
Question. Name two basic amino acids that provide positive charges to histone proteins.
Answer : Histone proteins have a positively charged surface because of the presence and abundance of two basic amino acids :
(i) Lysine and (ii) Arginine as compared to other amino acids.
Question. Which one of these an intron and an exon is reminiscent of antiquity ?
Answer : The primary transcript in eukaryotes contain both exons & introns (the non coding sequences). The introns are removed during processing by splicing & the exons are joined. The presence of introns in primary transcript is the reminiscent of antiquity.
Long Answer Type Questions
Question. (i) Describe the series of experiments of F. Griffith.
Comment on the significance of the results obtained.
(ii) State the contribution of Macleod, McCarty and Avery.
OR
(i) Explain with the help of Griffith’s experiment how the search for genetic material was conducted and what was the conclusion drawn ?
(ii) How did Macleod, McCarty and Avery establish the biochemical nature of the so called “genetic material” identified by Griffith in his experiment.
OR
(i) Describe the various steps of Griffith’s experiment that led to the conclusion of the ‘Transforming principle’.
(ii) How did the chemical nature of the ‘Transforming principle’ get established ?
OR
Describe Frederick Griffith’s experiment on Streptococcus pneumoniae. Discuss the conclusion he arrived at.
Answer : (i) Streptococcus pneumoniae :
S – Strain (virulent) injected into mice → mice die.
R – strain injected into mice → mice alive.
S – strain (heat killed) injected into mice → mice alive.
R – strain (alive) + S (heat killed) strain inject into mice → mice die.
R strain (non-virulent) picked up genetic material from S strain (virulent) and get transformed.
(ii) They (worked on the bio-chemical nature of transforming principle in Griffith’s experiment) purified proteins, DNA and RNA from heat killed S cells, they discovered protein digesting enzyme (protease), RNA digesting enzyme (RNAase) did not affect transformation, Digestion with DNase inhibited transformation, concluded DNA is the heredity material.
Detailed Answer :
(i) Griffith’s experiment : Griffith used mice and Streptococcus pneumoniae for his experiment.
Streptococcus pneumoniae has two strains :
(a) Smooth (S) strain (virulent) : It has polysaccharide mucus coat and can cause pneumonia.
(b) Rough (R) strain (Non-virulent) : It has no mucous coat and therefore do not cause pneumonia.
To test for the trait of pathogenicity, Griffith
injected mice with mixes of the two strains :
(c) S-strain → Inject into mice → Mice die
(d) R-strain → Inject into mice → Mice live
(e) S-strain (Heat killed) → Inject into mice → Mice live
(f) S-strain (Heat killed) + R-strain (live) → Inject into mice →Mice die
He concluded that some ’transforming principle’, transferred from heat-killed S-strain to R-strain. It enabled R-strain to synthesize smooth polysaccharide coat and become virulent. This must be due to the transfer of some genetic material.
(ii) Oswald Avery, Colin Macleod and McCarty worked to determine the biochemical nature of ‘transforming principle’ in Griffith’s experiment.
They purified biochemicals (proteins, DNA, RNA etc.) from the heat killed S cells to see which ones could transform live R cells into S cells.
They discovered that :
(i) Digestion of protein and RNA (using Proteases and RNases) did not affect transformation. So the transforming substance was not a protein or RNA.
(ii) Digestion of DNA with DNase inhibited transformation. It means that DNA caused transformation of R cells to S cells i.e. DNA was the transforming substance. Therefore they concluded that DNA is the hereditary material.
Question.

(i) Identify strands ‘A’ and ‘B’ in the diagram of transcription unit given above and write the basis on which you identified them.
(ii) State the functions of Sigma factor and Rho factor in the transcription process in a bacterium.
(iii) Write the functions of RNA polymerase-I and RNA polymerase-III in eukaryotes.
Answer : (i) A-Template strand
B-Coding strand
Template strand has polarity 3’-5’
Coding strand has polarity 5’-3’
On the basis of polarity with respect to promoter.
(ii) Sigma factor associates with RNA polymerase to initiate transcription, Rho factor gets associated to RNA polymerase to terminate transcription.
(iii) RNA polymerase I – Transcribes -RNAs
RNA polymerase III – Transcribes tRNA / 5srRNA / 5nRNA
Detailed Answer :
(i) Strand A is the template strand because it has 3’→5’ polarity. It acts as a template and is therefore so
called.
Strand B is called as the coding strand because it has 5’→3’ polarity. It does not get transcribed.
Template and coding strands are identified on the basis of polarity with respect to promoter.
(ii) In bacterium, the sigma (σ) factor initiates transcription. This σ factor recognizes the start signal and promotor region of the transcription unit which then along with RNA polymerase enzyme gets associated to the promotor to initiate transcription. Rho is the termination factor. It facilitate in terminating the process of transcription.
When the RNA polymerase reaches the terminator region the RNA polymerase is separated from DNA-RNA hybrid and as a result transcription terminates.
(iii) In eukaryotes, the RNA-Polymerase-I transcribes rRNAs (28S, 18S & 5S) & RNA-polymerase III transcribes tRNA, 5S rRNA & SnRNAs (Small nuclear RNAs).
In bacteria, a single DNA dependent RNApolymerase transcribes all the three types of RNAs i.e. mRNA, tRNA & rRNA).
Question. (i) Name the stage in the cell cycle where DNA replication occurs.
(ii) Explain the mechanism of DNA replication.
Highlight the role of enzymes in the process.
(iii) Why is DNA replication said to be semiconservative ?
Answer : (i) S-phase (synthesis phase) is the part of the cell cycle in which DNA replication takes place.
(ii) Process of DNA replication :
(a) The process of DNA replication begins at a point called the origin of replication (ori), to form a replication fork.
(b) The separated strands act as templates for the synthesis of new strands.
(c) DNA replicates in the 5’ → 3’ direction.
(d) dNTPs (Deoxyribonucleoside-tri phosphate) act as substrate and also provide energy for polymerization of nucleotides.
(e) DNA polymerase is an enzyme that assembles a new DNA strand that is complementary to the template strand.
(f) DNA polymerase continues to move along the template strand and add new nucleotides to the growing or complementary strand until the entire genome is replicated.
(g) The DNA polymerase forms one new strand (leading strand) in a continuous stretch in the 5’→3’ direction (Continuous synthesis).
(h) The other new strand is formed in small stretches (Okazaki fragments) in 5’ → 3’ direction (discontinuous synthesis).
(i) The Okazaki fragments are then joined together to form a new strand by an enzyme DNA ligase.
This new strand is called lagging strand.

(iii) DNA replication is said to be semi-conservative and discontinuous. Semi-conservative replication means that double stranded DNA molecule would produce two copies that each contained one of the original strands and one new strand.
Question. (i) How are the following formed and involved in DNA packaging in a nucleus of a cell ?
(a) Histone Octamer,
(b) Nucleosome,
(c) Chromatin.
(ii) Differentiate between Euchromatin and Heterochromatin.
Answer : (i) (a) Eight molecules of positively charged basic proteins called histones are organised to form histone octamer.
(b) Negatively charged DNA wrapped around positively charged histone octamer to give rise to nucleosome.
(c) Nucleosome constitute the repeating unit of a structure called chromatin.
(ii)

Detailed Answer :
(i) (a) Histone Octamer : It is a unit comprising two molecules each of four histones namely H2A, H2B, H3 and H4. These histone molecules are positively charged and interacts with negatively charged DNA to keep it associated with itself.
(b) Nucleosome : The negatively charged DNA wrapped around the histone octamer (positively charged) to form a nucleosome. Oudit (1975) called it as the nu-body. It consists of 200 base pairs and a histone octamer. Nucleosome act as building block for packaging the DNA into chromatin.
(c) Chromatin : It is thread like and a complex of DNA and histone protein. It helps in packing of DNA into the nucleus. The nucleosomes in chromatin look like beads on a string. The nucleosomes are the repeating units of chromatin and separated from one-another by the region of DNA which is coiled as linker DNA which is composed of 60 base pairs and one histone molecule H1. The nucleosomes are further coiled and packed into a structure of higher organization which is known as solenoid. Each solenoid is formed by six nucleosomes per turn.
Further supercoiling tends to form chromatin fiber and then chromatid. They further coils condense at metaphase stage to form chromosomes.
Question. (i) Describe the process of transcription in bacteria.
(ii) Explain the processing the hnRNA needs to undergo before becoming functional mRNA in eukaryotes.
OR
Explain the process of transcription in a prokaryote.
Answer : (i) Bacteria transcription is the process in which messenger RNA transcripts of genetic material in bacteria are produced to be translated for the production of proteins. Bacterial transcription occurs in the cytoplasm alongside translation.
The process of transcription is completed in three steps : Initiation, elongation and termination.
(a) Initiation : The enzyme binds at the promoter
site of DNA and initiates the process of transcription. It causes the local unwinding of the DNA double helix. An initiation sigma factor (σ) present in RNA polymerase initiates the RNA synthesis.
(b) Elongation : The RNA chain is synthesized in the 5’-3’ direction. RNA polymerase uses nucleoside triphosphate as substrate and polymerisation occurs according to complementarity.
(c) Termination : Termination occurs when termination factor (rho) alters the specificity of RNA polymerase to terminate the transcription.
As the RNA polymerase proceeds to perform elongation, a short stretch of RNA remains bound to the enzyme. As the enzyme reaches the termination region, this nascent RNA falls off and transcription is terminated.
(ii) The precursor of mRNA i.e. hnRNA contains
both introns and exons. Introns are removed and exons are joined by a process called splicing. The remaining mRNA is processed in two ways :
(a) Capping : Here, an unusual nucleotide called methyl guanosine triphosphate (cap) is added to the 5’ end of hnRNA.
(b) Tailing : Here, adenylate residues (200-300) are added at 3’ end of hnRNA in a template independent manner.
When hnRNA is fully processed, it is known as mRNA, which is transported out of the nucleus to get translated.
Question. How does the flow of information in HIV deviate from the Central Dogma proposed by Francis Crick.
Answer : According to Central Dogma of molecular biology, there is unidirectional flow of genetic information from DNA to m RNA and from here to protein.

But in HIV (Human immuno-deficiency virus) there is central dogma reverse i.e. the flow of genetic information is in reverse direction. It is because the virus contains the genetic RNA and produces an enzyme reverse transcriptase. This enzyme helps to synthesize DNA from genetic RNA of HIV by a process called as reverse transcription or Teminism.
This newly synthesized DNA functions as master copy producing mRNA through transcription & RNAs controlling translation to synthesize protein.
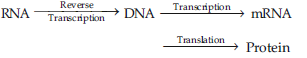
The phenomenon of reverse transcription was discovered by Temin & Baltimore (1970) in Retrovirus.
