Biotechnology Principles and Processes Notes for Class 12 Biology
Following are Biotechnology Principles and Processes Notes for Class 12 Biology. These revision notes have been prepared by expert teachers of Class 12 Biology as per the latest NCERT, CBSE, KVS books released for the current academic year. Students should go through Chater 1 Biotechnology Principles and Processes concepts and notes as these will help you to revise all important topics and help you to score more marks. We have provided Class 12 Biology notes for all chapters in your book. You can access it all free and download Pdf.
Chapter 11 Biotechnology Principles and Processes Notes Class 12 Biology
PRINCIPLES OF BIOTECHNOLOGY

TRADITIONAL HYBRIDISATION Vs GENETIC ENGINEERING
Traditional hybridization procedures very often lead to inclusion and multiplication of undesirable genes along with the desired genes.
Genetic engineering allows us to isolate and introduce only one or a set of desirable genes without introducing undesirable genes into the target organism.
GENETIC ENGINEERING INVOLVES
• Creation of recombinant DNA
• Use of gene cloning
• Gene transfer
The likely fate of a piece of DNA, which is somehow transferred into an alien organism :
➢ Most likely, this piece of DNA would not be able to multiply itself in the progeny cells of the organism
➢ But, when it gets integrated into the genome of the recipient, it may multiply and be inherited along with the host DNA. This is because the alien piece of DNA has become part of a chromosome, which has the ability to replicate. In a chromosome there is a specific DNA sequence called the origin of replication, which is responsible for initiating replication. Thus, an alien DNA is linked with the origin of replication, so that, this alien piece of DNA can replicate and multiply itself in the host organism. This can also be called as cloning or making multiple identical copies of any template DNA.
CONSTRUCTION OF THE FIRST ARTIFICIAL RECOMBINANT DNA MOLECULE
The construction of the first recombinant DNA was accomplished by Stanley Cohen and Herbert Boyer in 1972 .They adopted the following procedure.
• They isolated antibiotic resistance gene by cutting out a piece of DNA from the plasmid .
• The cutting of DNA at specific locations was done with the help of restriction enzymes popularly called as ‘molecular scissors.
• The cut piece of DNA with antibiotic resistance gene was then linked with the plasmid DNA of Salmonella typhimurium acting as vector with the help of the enzyme DNA ligase.
• This new autonomously replicating DNA created in vitro with linked fragment of antibiotic resistant gene is called recombinant DNA
• Recombinant DNA was then transferred into Escherichia coli, where it could replicate using the new host’s DNA polymerase enzyme. The ability to multiply copies of antibiotic resistance gene in E. coli was called cloning of antibiotic resistance gene in E. coli.

TOOLS OF RECOMBINANT DNA TECHNOLOGY :
The key tools are
• restriction enzymes
• polymerase enzymes
• ligases
• vectors
• the host organism
Restriction Enzymes:
• Restriction enzymes belong to a larger class of enzymes called nucleases.
• These are of two kinds; exonucleases and endonucleases.
Exonucleases remove nucleotides from the ends of the DNA whereas, endonucleases make cuts at specific positions within the DNA so that single stranded free ends called ‘sticky ends’ project from each fragment of DNA duplex.
• In 1963 two restriction enzymes responsible for restricting the growth of bacteriophage in Escherichia coli were isolated. One of these added methyl groups to DNA, while the other cut DNA. The later was called restriction endonuclease.
The first restriction endonuclease–Hind II was isolated and characterised in1968. It cuts DNA molecules at a particular point by recognising a specific sequence of six base pairs. This specific base sequence is known as the recognition sequence for Hind II.
Naming of restriction endonucleases is done as per the following conventions.
• First letter of the name comes from the genus of prokaryotic cell from which they were isolated. It is written in capital letters.
• Second two letters come from the species of the prokaryotic cell from which they were isolated. They are written in small letters.
• The fourth letter of the name represents the first letter of the strain.
• The Roman number written at the end of the name indicates the order in which the enzyme was isolated from that strain of prokaryotic cell.
e.g Naming of Restriction endonuclease EcoRI
• The capital letter comes from genus Escherichia of E. coli.
• Co comes from coli, the species of E. coli
• The letter R comes from RY 13 strain of E. coli.
• Roman number I indicates, it was the first enzyme isolated from bacterium E. coli.
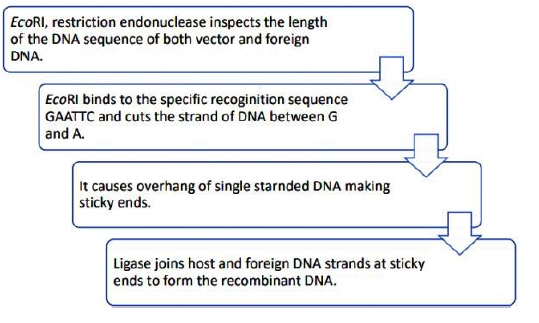
FUNCTIONING OF RESTRICTION ENDONUCLEASE
• Each restriction endonuclease functions by ‘inspecting’ the length of a DNA sequence.
• Once it finds its specific recognition sequence, it will bind to the DNA and cut each of the two strands of the double helix at specific points in their sugar-phosphate backbone.
• The recognition sequence of each restriction endonuclease consists of a definite number of specific base pairs in DNA double helix called palindromic nucleotide sequence. Palindromes are groups of letters that form the same words when read both forward and backward.
Steps in the formation of recombinant DNA by the action of restriction endonuclease enzyme –EcoR1

FLOW CHART SHOWING ACTION OF EcoR1:
SUMMARY OF RECOMBINANT DNA TECHNOLOGY(rDNA technology)

• -The restriction enzyme cuts both the vector DNA and the foreign DNA at the same site. It cuts each of the two strands of the DNA double helix at specific points in their sugar-phosphate backbone.
• This leaves overhanging sticky ends in foreign DNA and vector DNA.
• Sticky ends form hydrogen bonds with their complementary cut counterparts. This stickiness of the end facilitates the action of enzyme DNA ligase.
• -Thus Recombinant DNA is formed.
• The recombinant DNA is now shifted to the suitable host to get expressed and yield choice product.
Separation and isolation of DNA fragments: [GEL ELECTROPHORESIS]
-Gel electrophoresis is the technique used for separating DNA fragments.
PRINCIPLE OF GEL ELECTROPHORESIS:
DNA fragments are negatively charged. When an electric field is applied in a medium containing DNA, the DNA tend to move towards the positive electrode anode
PROCEDURE:
-The medium used in gel electrophoresis is agarose gel which is a natural polymer extracted from seaweeds.
-The DNA fragments are transferred to the agarose gel matrix and an electric field is applied.
-Under the electric field, the DNA fragments move according to their size through the seaving effect of agarose gel
-Smaller the size of the DNA fragment, the faster is the rate of movement of it and farther is the distance covered by it.
-The separated DNA fragments can be visualized by staining with ETHIDIUM BROMIDE followed by its exposure to UV radiations.
-The DNA appear in bright orange coloured bands in the gel
-The separated DNA bands can be cutout from the agarose gel and extracted from the gel. This step is known as ELUTION.

Cloning vectors
Cloning vectors are the DNA molecules that can carry a foreign DNA segment into the host cell.
(i) The vectors used in recombinant DNA technology can be:
(a) Plasmids Autonomously replicating circular extra-chromosomal DNA.
(b) Bacteriophages Viruses infecting bacteria.
(c) Cosmids Hybrid vectors derived from plasmids which contain cos site of X phage.
(ii) Copy number can be defined as the number of copies of vectors present in a cell.
(iii) Bacteriophages have high number per cell, so their copy number is also high in genome.
(iv) Plasmids have only one or two copies per cell.
(v) Copy number can vary from 1-100 or more than 100 copies per cell.
(vi) If an alien piece of DNA is linked with bacteriophage or plasmid DNA, its number can be multiplied equal to the copy number of the plasmid or bacteriophage.
Features Required to Facilitate Cloning into Vector
(a) Origin of replication (Ori)
(b) Selectable marker
(c) Cloning sites
(d) Vectors for cloning genes in plants and animals.
Origin of replication
Origin of replication(Ori) is a sequence from where replication starts.
• Any piece of DNA when linked to this sequence can be made to replicate within the host cells. The sequence is also responsible for controlling the copy number of the linked DNA.
(ii) Selectable marker helps in identifying and eliminating non-transformants and selectively permitting the growth of the transformants.
• Transformation is a process through which a piece of DNA is introduced in a host bacterium.
Selectable marker
• The genes encoding resistance to antibiotics such as ampicillin, chloramphenicol, tetracycline or kanamycin, etc, are some useful selectable markers for E. coli.
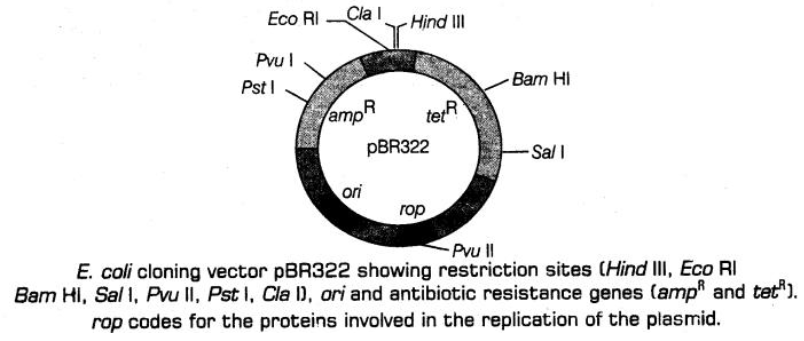
• Ligation of alien DNA is carried out at a restriction site present in one of the two antibiotic resistance genes. Example is ligating a foreign DNA at the Bam HI site of tetracycline resistance gene in the vector pBR322.
-» The recombinant plasmids will lose tetracycline resistance due to insertion of foreign DNA. But, it still can be selected out from non-recombinant ones by plating the transformants on ampicillin containing medium.
-» The transformants growing on ampicillin containing medium are then transferred on a medium containing tetracycline.
-» The recombinants will grow in ampicillin containing medium but not on that containing tetracycline.
The non-recombinants will grow on the medium containing both the antibiotics. In this example, one antibiotic resistance gene helps in selecting the transformants whereas, the other
antibiotic resistance gene gets ‘inactivated due to insertion’ of alien DNA and helps in selection of recombinants.
* Selection of recombinants due to inactivation of antibiotics is a cumbersome procedure, so alternative selectable markers are developed which differentiate recombinants from non-recombinants on the basis of their ability to produce colour in the presence of a chromogenic substrate.
-» In this method, a recombinant DNA is inserted within the coding sequence of an enzyme J3-galactosidase.
-» This results into inactivation of the enzyme, B-galactosidase (insertional inactivation).
-> The bacterial colonies whose plasmids do not have an insert, produce blue colour, but others do not produce any colour, when grown on a chromogenic substrate.
Cloning Site Cloning sites are required to link the alien DNA with the vector.
• The vector requires very few or single recognition sites for the commonly used restriction enzymes.
• The presence of more than one recognition sites within the vector will generate several fragments leading to complication in gene cloning.
(d) Vectors for cloning genes in plants and animals are many which are used to clone genes in plants and animals.
• In plants, the Tumour inducing (Ti) plasmid of Agrobacterium tumefaciens is used as a cloning vector.
-» Agrobacterium tumefaciens is a pathogen of several dicot plants.
-» It delivers a piece of DNA known as T-DNA in the Ti plasmid which 1 transforms normal plant cells into tumour cells to produce chemicals required by pathogens.
• Retrovirus, adenovirus, papillomavirus are also now used as cloning vectors in animals because of their ability to transform normal cells into cancerous cells.
Competent host organism
Competent host for transformation with recombinant DNA is required because DNA being a hydrophilic molecule, cannot pass through cell membranes, Hence, the bacteria should be made competent to accept the DNA molecules or Competency is the ability of a cell to take up foreign DNA.
Methods to make a cell competent are as follows.
(a) Chemical method in this method, the cell is treated with a specific concentration of | a divalent cation such as calcium to increase pore size in cell wall. The cells are then incubated with recombinant DNA on ice, followed by placing them briefly at 42°C and then putting it back on ice. This is called heat shock treatment.
• This enables the bacteria to take up the recombinant DNA.
(b) Physical methods In this method, a recombinant DNA is directly injected into the nucleus of an animal cell by microinjection method.
• In plants, cells are bombarded with high velocity microparticles of gold or tungsten coated with DNA called as biolistics or gene gun method.
(c) Disarmed pathogen vectors when allowed to infect the cell, transfer the recombinantDNA into the host.
ISOLATION OF THE GENETIC MATERIAL(DNA)
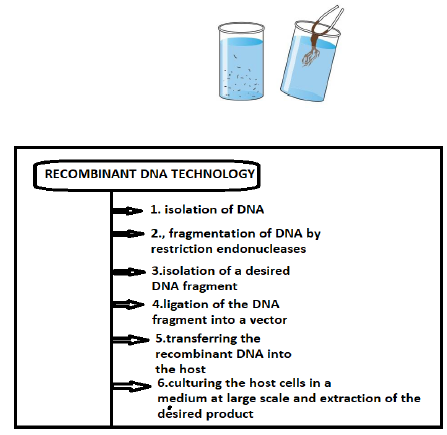
Genetic Material (DNA)
• RNA is removed by treatment with ribonuclease
• Proteins are removed by treatment with Protease
• The purified DNA is precipitated by adding chilled ethanol.
• The Bacterial/Plant /Animal cell is broken down by enzymes to release DNA, along with RNA,proteins ,polysaccharides and lipids.
• Bacterial cell is treated with enzyme Lysozyme
• Plant cell is treated with enzyme Cellulase.
• Fungal cell is treated with Chitinase.
Cutting DNA at specific locations
Using the restriction enzymes to cut target DNA and the vector DNA so that the sticky ends can be complimentary and linking can be done by ligase.
Amplification of Gene of Interest using PCR
Polymerase chain reaction (PCR) is the process in which the amplification of the gene of interest is carried out with two sets of primers and a thermostable DNA polymerase enzyme Taq polymerase. It was developed by Kary Mullis in 1985. The process involves the following steps:
Denaturation: The double-stranded DNA is heated up to 940 C which causes the hydrogen bonds to break and the two strands get separated.
Annealing: The two sets of primers are added which bind to the appropriate complementary segment of the DNA strand at 54oC.
Extension: The Taq polymerase enzyme polymerizes the nucleotide chain using the nucleotides provided in the medium and by using the template strand at 720 C.
Insertion of recombinant DNA into the host cell/organism.
There are several methods of introducing the ligated DNA into recipient cells.
Recipient cells after making them ‘competent’ to receive, take up DNA present in its surrounding.
If a recombinant DNA bearing gene for resistance to an antibiotic (e.g., ampicillin) is transferred into E. coli cells, the host cells become transformed into ampicillin-resistant cells.
Selectable marker-An antibiotic resistance marker is a gene that produces a protein that provides cells expressing this protein with resistance to an antibiotic.
Bacteria that have been subjected to a procedure to introduce foreign DNA are grown on a medium containing an antibiotic, and those bacterial colonies that can grow have successfully taken up and expressed the introduced genetic material.
Normally the genes encoding resistance to antibiotics such as ampicillin, chloramphenicol, tetracycline or kanamycin, etc., are considered useful selectable markers.
Obtaining the foreign gene product
In recombinant technologies, the desired genes for administration is selected, followed by selecting a perfect vector into which the desired gene has to be integrated and recombinant DNA formed.
Once this foreign DNA is inserted, it is multiplied and ultimately desirable protein is produced.
And at last, it has to be maintained in the host and carried forward to the offspring.
For the production of the desired protein, the gene encodes for it needs to be expressed.
This happens only under optimized conditions.
The target protein has to be expressed and produced on a large scale.
The recombinant cells can be multiplied in large scale using a continuous culture system.
Here the cells are cultured in a large vessel and the medium is refreshed on a regular interval to maintain the optimum conditions.
This helps to culture a large mass of the desired protein. This was achieved by the development of bioreactor.
Recombinant protein is the protein encoding gene expressed in a heterologous host
Bioreactors are vessels in which raw materials are biologically converted into specific products using microbial, plant or animal cells.
Bioreactor is designed to meet several requirements such as pH and temperature control, aeration and agitation, drain or overflow and sampling facility.
Regular monitoring for physical, chemical and biological parameters is done through control systems of the bioreactor.
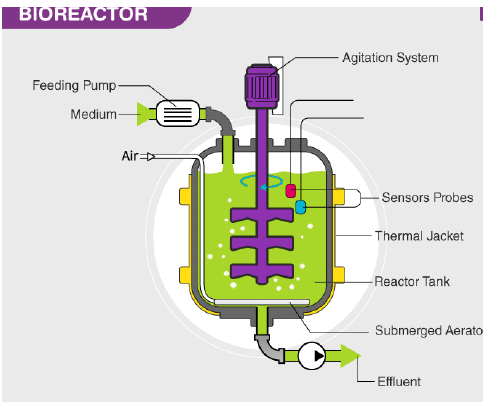
TYPES OF BIOREACTOR:
(a) Simple stirred-tank bioreactor;
(b) Sparged stirred-tank bioreactor.
Downstream Processing.
Downstream Processing: The process of formulation, separation and purification of rDNA products made in Bioreactors.
Downstream processing is a sequential step in which the isolation, purification, preservation of final products are done before it is marketed.
In this stage, the final product is formulated with additives like preservatives, colors, etc, followed by clinical trials.
Strict quality control testing for each product is also required
Important Questions Biotechnology Principles and Processes Class 12 Biology
Question. List the major techniques in Genetic Engineering? Who was the first to construct an rDNA?
Answer : Creation of rDNA, gene cloning ,gene transfer, Stanley Cohen & Boyer
Question. How does genetic engineering help in overcoming the limitations of traditional hybridization procedures used in plants & animals?
Answer : It allows use to isolate & introduce only the desirable gene/genes without Introducing the undesirable genes.
Question. Show diagrammatically how end nucleases work?
Answer : Fig 11.1
Question. How can an alien piece of DNA made to multiply in a host cell?
Answer : Linked to ori of the host genome.
Question. Explain the features required for a cloning vector
Answer : ori, cloning sites, selectable markers
Question. What are the three basic steps in genetically modifying an organism?
Answer : Identification of desirable DNA, introduction into host, maintenance and then transfer to its progeny.
Question. What are the two core techniques that enabled the birth of Biotechnology?
Answer : Genetic Engineering & Maintenance of sterile ambience.
Question. What are recognition sequences often do nucleases? Name the five key tools in rDNA technology.
Answer : The sequence at which DNA is cut by a Restriction Endonuclease polymerases, ligase, vector, host
Question. What are nucleases. What are the two types?
Answer : Enzymes that act on nuleic acids, Endonucleases ,Exnucleases.
QuestionSequentially state the process you would adopt for getting a recombinant protein?
Answer : Isolation of DNA(enzymes used),cutting the DNA, separation of fragments , PCR, introducing into host cell, obtaining gene product in the Bioreactor,downstream processing.
Question. How does the stickiness of cut ends of DNA help?
Answer : Makes the joining easy ; Complimentary.
Question. How can the fragments of DNA be separated? Explain
Answer : Gel electrophoresis-Explain Agarose gel
Question. What are the ‘molecular scissors’ in rDNA technology. What are they used for in rDNA technology?
Answer : Restriction enzymes, DNA at specific sites
Question. How can the cloning vector pBR322 be used in separating the transform ants and recombinants? .Explain
Answer : Tetracycline site can be cut ,to insert the desired insert ,the recombinants will lose resistance to tetracycline the transform ants will have resistance to both tetracycline and ampicillin
Question. How is Agra bacterium tumifaciens used in rDNA technology. Explain ii)What and how are other pathogens are used for the purpose ?State two other methods by which host organism can be transformed?
Answer : Used as vector ,by modifying tumour inducing Ti plasmid, it is not pathogenic, transforms the host plant cell Retrovirus, disarmed
